Dear experts,
We are relatively new to Corryvreckan and are running into issues with the alignment procedure for our test beam setup.
The setup consists of six MIMOSA 26 telescope planes and a single 1.3x1.3 mm2 LGAD sensor in the middle, i.e. three MIMOSA planes on either side of the sensor. Instead of actual test beam data, we are using simulated data generated by Allpix2 for a 5 GeV square beam of electrons (uniformly distributed) with sides of 1.35 mm which we load in via the FileReader module.
Issue regarding the alignment of the telescope
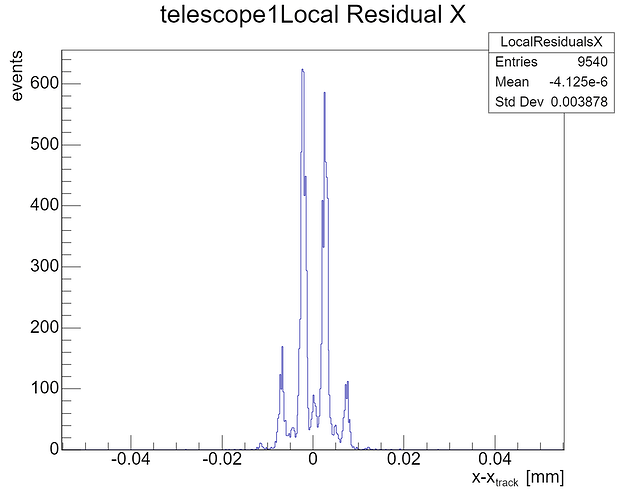
After one iteration of the Prealignment module for the telescope and three iterations of AlignmentTrackChi2 (decreasing the spatial_cut_abs for each iteration from 200 um, to 100 um, to 50 um), the residuals of telescope1 and telescope4 – the ones that are in the middle for the triplet of MIMOSA planes before and after the DUT – still display a ‘spiky’ pattern with roughly evenly spaced peaks. (See the picture below and the configs in the attachment.)
We were wondering if someone could explain
a) the cause of these patterns in the residual plots to us;
b) why they are only present in the plots for telescope plane 1 and 4; and
c) if this is something to worry about.
We have tried using different track reconstruction models (gbl is used for the image above, but we have also tried straightline), but the peaks show up regardless.
Issue regarding the alignment of the DUT
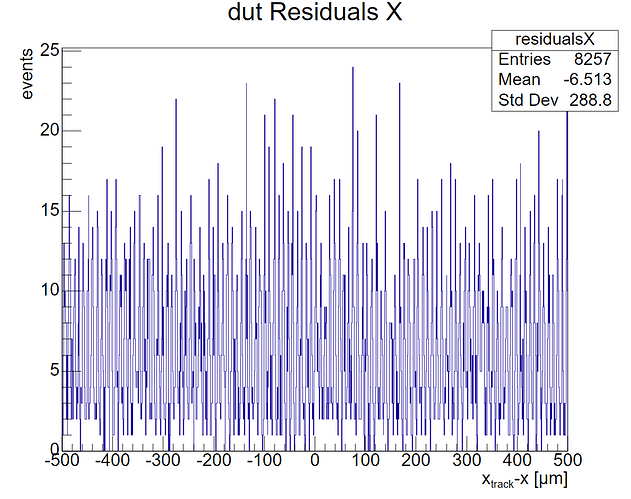
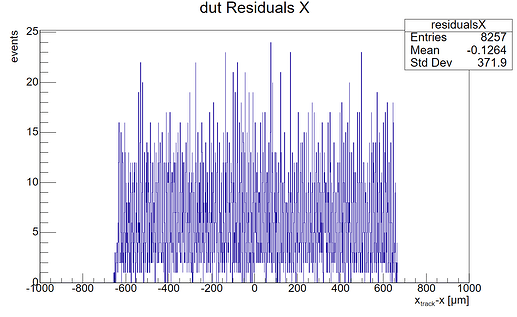
For aligning the DUT, we adopted a similar procedure using AlignmentDUTResidual and decreasing the spatial_cut_abs from 5 mm to 2.5 mm to 1.5 mm. The issue here is that we have no way of validating the alignment because the residuals, as expected, display a uniform distribution. Since our DUT only consists of one sensor and all clusters are reconstructed in the centre, the residuals simply show the distance between the interpolated track intercepts – that are uniformly distributed over the sensor - and the middle of the sensor.
Our question is: is there an alternative way to validate the alignment of the DUT? We know for a fact that it is currently off because the position of the DUT provided by Corryvreckan differs about 8 um from the position of the DUT we have simulated our data with. Hence our interest in improving the alignment.
Thanks a lot in advance,
Robin & Andrea
DUT_alignment.conf (909 Bytes)
telescope_alignment.conf (884 Bytes)